Abstract
Multiple myeloma (MM) is a malignancy of terminally differentiated plasma cells (PCs) that primarily grow in the bone marrow (BM). MM arises from the pre-malignant condition monoclonal gammopathy of undetermined significance (MGUS), in which the accumulation of chromosomal copy number variations (CNVs), chromosomal rearrangements, and somatic mutations leads to the expansion of a clonal PC population. CNVs and translocations present at time of diagnosis play a significant role in patient prognosis and in the stratification of high-risk multiple myeloma (hrMM). At the level of gene expression, two independent gene signatures predicting increased relapse risk and poorer overall survival have been developed for use in diagnosing hrMM: the UAMS-70 and EMC-92 gene signatures.
Recently, our lab developed the first genetically engineered NrasQ61R; Myc-driven myeloma model (VQ myeloma). This so-called VQ model shows characteristics representative of hrMM. In our initial study, five lines of VQ myeloma were isolated from primary mice (VQ-D1 through VQ-D5). In four of these VQ lines (VQ-D1, D2, D4, and D5), we established survival of recipient mice transplanted with donor VQ cells and carried out bulk RNA-Seq analysis. Finally, we established two VQ cell lines from two VQ-D2 recipients, so-called VQ 4935 and 4938.
Although all VQ cells are driven by MYC dysregulation and RAS hyperactivation, different lines display distinct patterns of extramedullary disease, antibody sub-type, and survival. However, the genetic and/or transcriptional bases for these phenotypic differences remain elusive. Additionally, what subtypes of hrMM they may individually represent (if any) are unknown. In the current study, we aim to address these questions through the molecular characterization of five primary VQ lines using B cell receptor (BCR) heavy chain repertoire sequencing, whole exome sequencing, copy number variation analysis, and RNA-Seq.
Results showed BCR amplicons that were either clonal or oligoclonal with a dominant clone present in the four primary VQ lines. With the exception of the VQ 4935 cell line, BCR repertoire analysis showed relatively low rates of somatic hypermutation (SHM) in VQ cells compared to human myeloma, with mutation frequencies ≤3.0%. Although median SHM frequency was slightly higher in Cluster I VQ (VQ-D2/D5), there was no significant correlation with median survival of different VQ lines.
WES of five primary VQ lines identified 11 recurrently mutated genes. Human orthologs of two of these genes, FAT4 and SP140, have been identified as recurrently mutated in numerous MM patient sequencing studies. STRING and RNAseq analyses suggest that Fat4 serves a passenger role in MM progression, while Sp140 plays a role in interferon signaling in VQ cells.
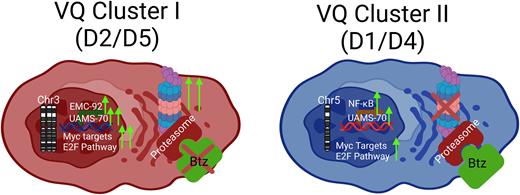
We performed CNV analysis in five VQ lines and identified two recurrent, mutually exclusive events: amplification of chr3, which was present in Cluster I (VQ-D2, D3, and D5), and monosomy chr5, which was present in Cluster II (VQ-D1 and D4). CNVs identified in the VQ model include several tumor suppressor genes and oncogenes that are highly relevant for MM pathogenesis. Notably, chr3 includes the proto-oncogene Nras and the tumor suppressor Fam46c, while Chr5 includes Cdk6, Fgfr3, and Mmset. Based on RNAseq analysis, we found that gene copy numbers do not necessarily correlate with mRNA levels in VQ cells, suggesting that additional epigenetic mechanisms play an important role in controlling gene expression.
Transcriptional analysis of RNAseq data via unsupervised hierarchical clustering, tSNE and PCA analyses revealed that VQ myeloma lines cluster into two distinct transcriptional subtypes corresponding to the presence of recurrent CNVs. Extensive pathway analysis showed that Cluster I VQ was enriched for cancer growth pathways and hrMM gene signatures compared to Cluster II and t-VĸMyc (Vk12653), while Cluster II was enriched for NF-ĸB signaling and overexpressed genes associated with bortezomib (Btz) response in human MM. Consequently, in sharp contrast to Cluster II VQ cells that showed short-term response to Btz, Cluster I VQ cells were de novo resistant to Btz in vivo. Our study provides insights into the heterogeneity of VQ lines and highlights Cluster I VQ lines as highly representative of human hrMM.
Disclosures
Patnaik:Kura Oncology, Stem Line Pharmaceuticals: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal